The Model
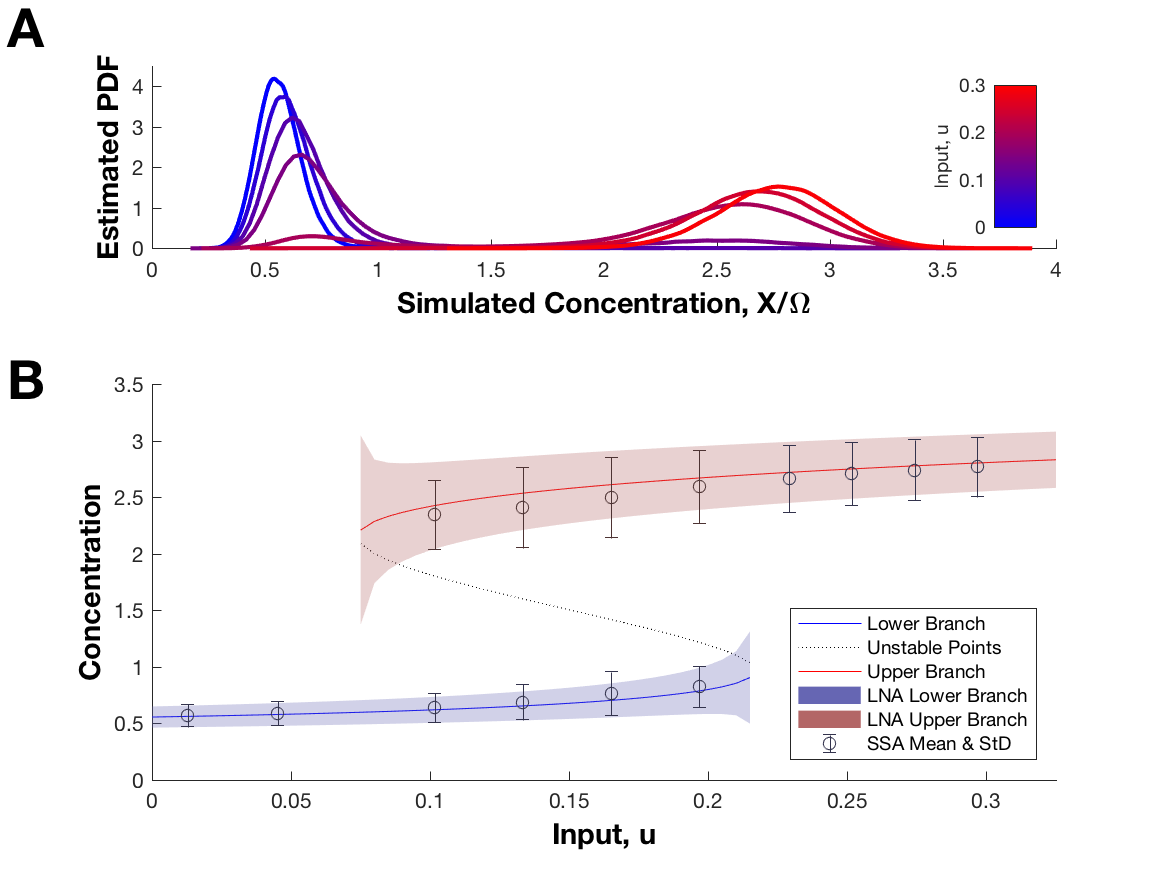
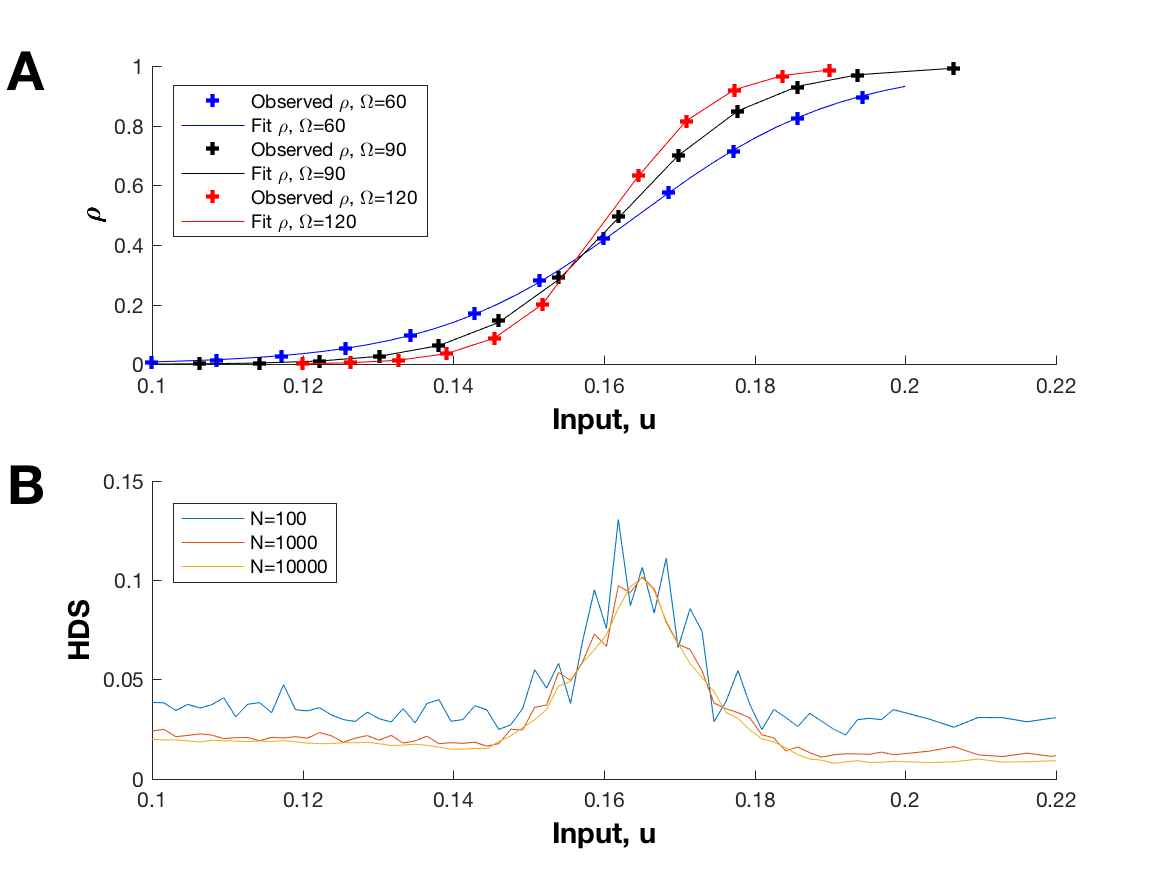
Bistable gene-expression systems are the equivalent of a toggle switch in synthetic biology and therefore may function as a component in performing logical operations. In practice some switching mechanisms can be controlled by varying some continuous (experimentally controllable) parameters. However, when attempting to model bistable systems, the parameter estimates obtained via the standard fitting procedure are often of low accuracy due to the variability of experiments. This effect is pronounced in unoptimized experiments. Optimal experimental design procedures are used to select the experiment which best reduces parameter estimation error.
This is a quick demo of the stochastic gene expression model that I studied during one of my undergraduate research projects at the University of Waterloo. I helped our group by implementing the Gillespie algorithm for this model, using this method to generate a lot of data, and performing some statistical analysis. For more information, please see our conference paper [1]. The input control variable, u, can be adjusted in the simulation to demonstrate bistable switching.
Data